Rapid CE microbial assays
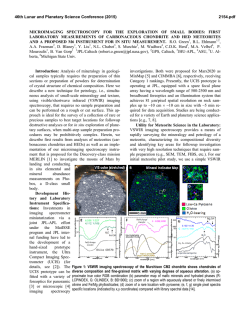
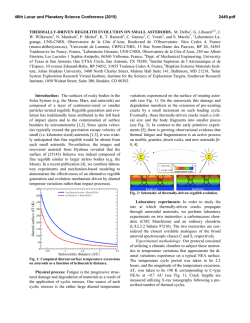
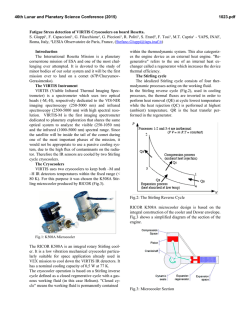
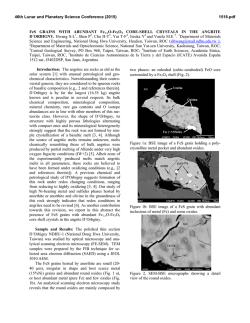
FEMS Microbiology Letters 194 (2001) 33^37 www.fems-microbiology.org Rapid CE microbial assays for consumer products that contain active bacteria Daniel W. Armstrong *, Je¡rey M. Schneiderheinze, John P. Kullman, Lingfeng He Department of Chemistry, Iowa State University, Ames, IA 50011, USA Received 7 August 2000 ; received in revised form 16 September 2000; accepted 22 September 2000 Abstract Recent advances in high efficiency separation methods of bacteria allow their rapid identification and quantitation in some cases. A specific capillary electrophoresis (CE) technique is used to identify and quantitate Lactobacillus acidophilus in both pill and syrup health products as well as Bifidobacterium infantis in a powdered formula supplement. Cell viability can be evaluated as well. In some cases, both the living and dead bacterial cells as well as the molecular excipients can be evaluated in a single run. ß 2001 Federation of European Microbiological Societies. Published by Elsevier Science B.V. All rights reserved. Keywords : Capillary electrophoresis ; Cell viability ; L. acidophilus; B. infantis 1. Introduction There are increasing numbers of health aids, supplements and consumer products in which the `active ingredient' is not a compound, but rather a microbe. Many reports indicate that speci¢c microbes can be bene¢cial to human health. Often this is due to the microbe's ability to produce needed or useful substances [1]. In other instances a microbe may degrade or alter unwanted or detrimental substances [2^7]. Also it is known that a healthy population of benign microorganisms can inhibit or retard the growth of unwanted or pathological (in human terms) species [1,2]. Some of the most prevalent examples of products in which the active ingredient is a bacterium, are pills and syrups for individuals that are lactose intolerant. It is estimated that approximately 50 million Americans su¡er from lactose malabsorption, which produces a number of clinical symptoms [4]. The purity, activity and e¡ectiveness of the available products for treating lactose maldigestion are open to question [2^7]. One of the problems is that there are no e¡ective assays for the active bacterial ingredient(s). This is important since it is known that both the viability and the nature (species and strain) of the * Corresponding author. Tel. : +1 (515) 294-1394; Fax: +1 (515) 294-0838; E-mail: [email protected] bacteria determine whether or not it produces any bene¢cial e¡ects [1,2,5,7]. There are numerous other examples of commercial bacterial preparations for which there are no fast, e¤cient or sensitive assays. This includes medicinal, food and other consumer products. Recently it has been demonstrated that speci¢c capillary electrokinetic techniques can be used to rapidly separate, identify and quantitate bacteria and fungi [8^11]. If it is possible to adapt these methods so that microbes can be assayed in complex consumer products (in much the same way as molecules) it would be extremely useful. In this communication we report the ¢rst high e¤ciency microbial assays of tablets (pills) and powder-based commercial products. 2. Materials and methods 2.1. Materials Tris(hydroxymethyl)aminomethane (TRIS), boric acid, ethylenediaminetetraacetate (EDTA), and poly(ethylene) oxide (PEO; Mn = 600 000) were purchased from Aldrich (Milwaukee, WI, USA). Milk-free Acidophilus distributed by Schi¡ Products, Inc. (Salt Lake City, UT, USA) and Spring Valley brand natural Acidophilus manufactured by Rexall Sundown, Inc. (Boca Raton, FL, USA) are dietary supplements containing Lactobacillus acidophilus. Solaray brand BabyLife1 dietary supplement manufactured by 0378-1097 / 01 / $20.00 ß 2001 Federation of European Microbiological Societies. Published by Elsevier Science B.V. All rights reserved. PII: S 0 3 7 8 - 1 0 9 7 ( 0 0 ) 0 0 5 0 2 - 4 FEMSLE 9719 21-12-00 34 D.W. Armstrong et al. / FEMS Microbiology Letters 194 (2001) 33^37 Nutraceutical Corp. for Solaray, Inc. (Park City, UT, USA) contains Bi¢dobacterium infantis. All three dietary supplements were purchased from a local nutrition store. A LIVE/DEAD BacLight Bacterial Viability Kit was purchased from Molecular Probes, Inc. (Eugene, OR, USA). It consists of SYTO 9 green £uorescent nucleic acid stain and red £uorescent nucleic acid stain, propidium iodide, which are dissolved separately in DMSO. 2.2. Methods A stock bu¡er solution containing 4.5 mM TRIS, 4.5 mM boric acid and 0.1 mM EDTA (TBE bu¡er) was prepared by dissolving appropriate amounts of each reagent in deionized water yielding a bu¡er of pH 8.4. This bu¡er solution was then diluted 8:1 with deionized water (diluted TBE bu¡er). A stock polymer solution was prepared by dissolving 0.2 g of PEO in 40 ml of the diluted TBE bu¡er. This solution was sonicated for 2^3 h at 50³C (Fisher model FS-28, 720 W at 43 kHz) to facilitate the dissolving process. The mixture was then removed from the bath and left to completely dissolve by stirring overnight at room temperature. The CE running bu¡er was prepared by diluting the stock polymer solution with the diluted TBE bu¡er to a ¢nal polymer concentration of 0.025% (v/v). All bu¡ers and polymer solutions were prepared fresh daily. Cultures were grown in-house from the three dietary supplements. Samples were added to Nutrient Broth (Difco Laboratories, Franklin Lakes, NJ, USA) and were grown for 10^24 h at 30³C on a shaker at 250 rpm. ware. Fused silica capillary with a 100 Wm i.d. was purchased from Polymicro Technologies, Inc. (Phoenix, AZ, USA). The column used for the separations was 27 cm in length (20 cm to window). Prior to each injection, the column was washed for 0.5 min with water, 1.5 min with 1 N NaOH, 0.5 min with water, followed by 0.5 min with the running bu¡er. The samples were pressured injected for 10^12 s at 0.5 psi. The separation was performed at a voltage of 10 kV and a temperature of 25³C with thermostated control. On-line UV detection of the samples was accomplished at 214 nm. The detector response results from a combination of light scattering and absorbance. 2.4. Quantitation of bacteria Samples were prepared for quantitation by dissolving 1, 1.5, 2, 3 and 4 Schi¡ tablets in 20 ml of the CE running bu¡er. Samples were then injected into the P/ACE 5000 CE system for 20 s at 0.5 psi. The separation was performed at voltage of 10 kV and a temperature of 25³C. On-line detection was accomplished at 214 nm. The relative standard deviation (RSD) of the peak areas was calculated to be 2.85% (n = 3). 2.3. Capillary electrophoresis The pill samples were prepared by dissolving the tablets in 10 ml of CE running bu¡er in a 40 ml sample vial. The sample was shaken by hand and allowed to settle for 20^ 30 min under the in£uence of gravity. This allowed the larger insoluble pieces of the pill to settle to the bottom of the sample vial leaving a cloudy solution above the solid particles. 2^3 ml of cloudy solution was then taken as the sample. Visual microscopy (400U) con¢rmed the presence of bacteria in the samples and also pieces of unidenti¢ed insoluble particles. The sample removed from the 40 ml vial was then used for CE analysis. Samples from the bacterial cultures grown from the tablets were obtained by removing 6^7 ml of liquid culture and pelleting the cells in a centrifuge (Fisher model 228, Pittsburgh, PA, USA) at 3400 rpm. The supernatant was decanted and the cells were washed with 1^2 ml of CE running bu¡er and again pelleted in the centrifuge. After another wash, the cells were dispersed in 1^2 ml of CE running bu¡er and used for analysis. The CE separations were performed on a Beckman P/ ACE 2100 or P/ACE 5000 (Palo Alto, CA, USA) coupled to a computer equipped with Gold data acquisition soft- Fig. 1. Electropherograms of (A) cultured cells of B. infantis from Babylife powder, and (B) direct injection analysis of dissolved Babylife powder. See Section 2 for experimental details. FEMSLE 9719 21-12-00 D.W. Armstrong et al. / FEMS Microbiology Letters 194 (2001) 33^37 35 equipped with a 520 nm bandpass ¢lter and a 663 nm longpass ¢lter, respectively. Data were collected with P/ACE system MDQ software. The ratio of green £uorescence peak area to the red £uorescence peak area can be correlated to the ratio of live cells to dead cells in a pill after correcting for spectral overlap and normalizing the peak areas to known concentrations of live and dead cells. Relatively high concentrations of the dyes are used (compared to biological staining procedures) in order to maximize staining of the cells and produce consistent results. The CE-LIF cell viability results were con¢rmed by £ow cytometry using a Beckman Coulter EPICS XL-MCL instrument. 3. Results and discussion Speci¢c electrokinetic separation conditions can be engineered so that microbes of the same type elute as single peaks [8,9]. This should allow the development of rapid instrumental assays and quantitation for microorganisms. While high e¤ciency separation-based assays are common for molecules, ions, etc., nothing comparable has ever been demonstrated for microorganisms. Fig. 2. Electropherograms of (A) cultured cells of L. acidophilus from Schi¡ tablets, and (B) direct injection analysis of dissolved Schi¡ tablets. See Section 2 for experimental details. 2.5. Viability determination Samples were prepared by dissolving one Schi¡ tablet in 20 ml CE running bu¡er. Three ml aliquots of cloudy solution were taken as samples. Approximately 1 Wl of 20 mM propidium iodide and 10 Wl 3.34 WM SYTO 9 solution was added to a 3 ml sample solution. The sample was incubated in the dark for 30 min. Generally, SYTO 9 stain labels all bacteria in a population ^ those with intact membranes and those with damaged membranes. In contrast, propidium iodide penetrates only bacteria with damaged membranes, causing a reduction in the SYTO 9 stain £uorescence when both dyes are present. Thus, with an appropriate mixture of SYTO 9 and propidium iodide stains, live bacteria with intact cell membranes stain £uorescent green, whereas dead bacteria with damaged membranes stain £uorescent red. Viability determination was completed on a Beckman Coulter P/ACE MDQ capillary electrophoresis system, equipped with 488 nm laser induced £uorescence (LIF) detector. The sample was injected for 5^10 s at 0.5 psi. The separations were performed at 15 kV and a temperature of 25³C. Green and red £uorescent light were monitored simultaneously with the LIF detector, which is Fig. 3. Electropherograms of (A) cultured cells of L. acidophilus from Spring Valley tablets, and (B) direct injection analysis of dissolved Spring Valley tablets. See Section 2 for experimental details. FEMSLE 9719 21-12-00 36 D.W. Armstrong et al. / FEMS Microbiology Letters 194 (2001) 33^37 Fig. 4. Plot showing the correlation between peak area and the number of L. acidophilus cells in injected samples from Schi¡ pills. Such standard curves (y = 0.046^2.51, R2 = 0.995) allow rapid and accurate quantitation of bacteria. An electropherogram for a culture grown standard of B. infantis is shown in Fig. 1A. These bacteria are in certain products (see Section 2) for introduction to the guts of newborn infants. The powdered formulation is a relatively simple mixture of maltodextrin and the bacterium B. infantis. The CE electropherogram of this formulation shows that both the bacteria and the maltodextrin (which travels with the eof) can be identi¢ed and quantitated in the same run (Fig. 1B). Several formulations contain L. acidophilus as the active ingredient. An assay of L. acidophilus from a commercial syrup (Fig. 2B) shows that the L. acidophilus (as identi¢ed from the standard in Fig. 2A) as well as at least two of the molecular components of this preparation can be identi¢ed. Note that the conditions of these assays (i.e. the pH, strength, bu¡er type and polymer concentration of the running bu¡er) can usually be controlled so that the larger, charged microbes elute latter than most of the excipients. Finally L. acidophilus was analyzed from a complex tablet formulation that contained six or more molecular or colloidal excipients. The electropherograms of the bacteria standard and a solution of the tablet are shown in Fig. 3A and B respectively. The purpose of this ¢gure is two-fold. First, it shows that a bacteria can be assayed in widely di¡erent formulations (i.e. matrices). Second, mi- gration times can vary slightly (see L. acidophilus in Fig. 3A vs. B) in the presence of certain matrices. Thus an internal standard or sample spiking should be used to verify one's results (at least initially). Most often a component or components of the injected sample change the eof, which then a¡ects the migration of other components in that run. This same phenomenon was noted recently when the direct injection of concentrated urine was used for the CE diagnosis of urinary tract infections [9]. The CE peak area (Fig. 4) can be directly correlated to the number of cells injected (or the weight% of cells injected). Thus, the direct and rapid quantitation of cells in a sample or environment can be done. The ability to both e¤ciently identify and quantitate microorganisms can be di¤cult or impossible using conventional or classical methods. Electropherograms generated using UV detection (Figs. 1^3) allow identi¢cation of a microorganism and quantitation of the total number of cells (Fig. 4). However, they give no information on cell viability. Cell viability can be determined in the same CE run by pretreating the cells with a mixture of SYTO 9 green £uorescent nucleic acid stain plus red £uorescent propidium iodide stain and using dual channel, laser induced £uorescence detection (see Section 2). Viable cells produce an enhanced green £uorescence while the nonviable cells produce a red £uorescence. FEMSLE 9719 21-12-00 D.W. Armstrong et al. / FEMS Microbiology Letters 194 (2001) 33^37 37 determine microbe viability in a single run is a great advantage. It is expected that applications of this sort will expand in the future, not only in research laboratories but also in regulatory agencies and bio-industrial areas. References Fig. 5. CE/LIF electropherograms generated by simultaneously monitoring the (A) green £uorescence of viable cells (520 nm) and the (B) red £uorescence of the dead cells ( s 662 nm) obtained from Schi¡ tablets. The uncorrected peak areas are shown in parentheses in each ¢gure. The corrected relative peak areas show that approximately 40% of the L. acidophilus in this sample was nonviable. See Section 2 for experimental details. Under appropriate conditions and simultaneously monitoring the relative £uorescence intensities (Fig. 5), the ratio of viable to nonviable cells can be obtained [11]. Analysis shows that only about 60% of the L. acidophilus cells in the Schi¡ tablet samples are viable (Fig. 5). The application of high e¤ciency microbial separations (HEMS) to commercial products has been demonstrated for the ¢rst time. The ability to identify, quantitate, and [1] Ehrlich, H.L. (1985) In: Bacteria in Nature, Vol. 1, Bacterial Activities in Perspective (Leadbetter, E.R. and Poindexter, J.S., Eds.), Ch. 7, pp. 189^219. Plenum Press, New York. [2] Mitsuoka, T. (1978) Intestinal Bacteria and Health. Harcourt Brace Jovanovich, Tokyo. [3] Mustapha, A., Jiong, T. and Savaiano, D.A. (1997) Improvement of lactose digestion by humans following ingestion of unfermented Acidophilus milk: In£uence of bile sensitivity, lactose transport and acid tolerance of Lactobacillus acidophilus. J. Dairy Sci. 80, 1537^ 1545. [4] Suarez, F.L., Savaiano, D.A. and Levitt, M.D. (1995) Review article: The treatment of lactose intolerance. Aliment Pharmacol. Ther. 9, 589^597. [5] Vesa, T.H., Marteau, P., Zidi, S., Briet, F., Pochart, P. and Rambaud, J.C. (1996) Digestion and tolerance of lactose from yogurt and di¡erent semi-solid fermented dairy products containing Lactobacillus acidophilus and Bi¢dobacteria in lactose maldigesters : Is bacterial lactose important? Eur. J. Clin. Nutr. 50, 730^733. [6] Lin, M.-Y., Yen, C.-L. and Chen, S.-H. (1998) Management of lactose maldigestion by consuming milk containing lactobacilli. Dig. Dis. Sci. 43, 133^137. [7] Martini, M.C., Lerebours, E.C., Lin, W.J., Harlander, S.K., Berrada, N.M., Antoine, J.M. and Savaiano, D.A. (1991) Strains and species of lactic acid bacteria in fermented milks yogurts e¡ect on in-vivo lactose digestion. Am. J. Clin. Nutr. 54, 1041^1046. [8] Armstrong, D.W., Schulte, G., Schneiderheinze, J.M. and Westenberg, D.J. (1999) Separating microbes in the manner of molecules I. Capillary electrokinetic approaches. Anal. Chem. 71, 5465^5469. [9] Armstrong, D.W. and Schneiderheinze, J.M. (2000) Rapid identi¢cation of bacterial pathogens responsible for urinary tract infection using direct injection CE. Anal. Chem. 72, 4474^4476. [10] Schneiderhinze, J.M., Armstrong, D.W., Schulte, G. and Westenberg, D.J. (2000) High e¤ciency separation of microbial aggregates using capillary electrophoresis. FEMS Microbiol. Lett. 189, 39^44. [11] Armstrong, D.W. (2000) Patents pending. FEMSLE 9719 21-12-00
© Copyright 2026