Clinical Utility: Informing Treatment Decisions by Changing the
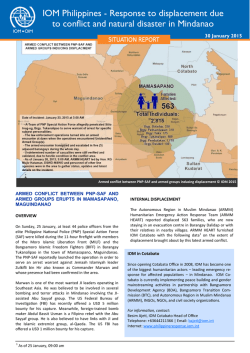
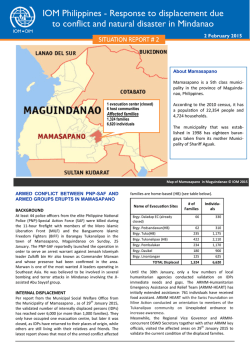
Clinical Utility: Informing Treatment Decisions by Changing the Paradigm Robert T. McCormack and Paul R. Billings* January 29, 2015 *The authors are participants in the activities of the IOM Roundtable on Translating GenomicBased Research for Health. The views expressed in this discussion paper are those of the authors and not necessarily of the authors’ organizations or of the Institute of Medicine. The paper is intended to help inform and stimulate discussion. It has not been subjected to the review procedures of the Institute of Medicine and is not a report of the Institute of Medicine or of the National Research Council. Copyright 2015 by the National Academy of Sciences. All rights reserved. The types of evidence needed to support the use of genome sequencing in the clinic varies by stakeholder and circumstance. In this IOM series, seven individually authored commentaries explore this important issue, discussing the challenges involved in and opportunities for moving clinical sequencing forward appropriately and effectively. Clinical Utility: Informing Treatment Decisions by Changing the Paradigm Robert T. McCormack, Janssen Oncology; and Paul R. Billings, Thermo Fisher Scientific1, 2 Recent improvements in cancer treatment and patient survival can be attributed, in part, to several initiatives. Most notable was the challenge by the Food and Drug Administration (FDA) to pharmaceutical and device developers to identify biomarkers that determine those patients most likely to respond to a drug. This has long been considered an essential step toward personalized medicine. Achieving this challenge was greatly enabled by the substantial technical innovations that made it possible to identify these new markers. The development of nextgeneration sequencing, for instance, provided a groundswell of information on genetic changes that drive the growth of tumors, and the technology continues to form the basis for development of these highly specific and effective drugs. Today, more than 100 molecularly defined biomarkers are associated with commercially available drugs or drugs under development (Aston and Lovly, 2014). Our success in developing targeted therapies has been impressive, but the path forward is challenging and filled with uncertainty. For example, next-generation sequencing and other newly developed technologies that work sequentially or in parallel to sequencing technologies have enabled nearly complete analysis of a patient’s DNA (genome), RNA (transcriptome), protein (proteome), and posttranslational (epigenomic) modifications, yielding significant insights into the basic biology of cancer. However, these technologies have also brought to light the true heterogeneity or complexity of common cancers and revealed that tumor resistance to therapy occurs frequently, underscoring the need to continually assess tumors with these technologies to anticipate tumor behavior and retarget therapy. All this information is directional toward developing new anti-cancer treatments, but the data need to be considered with the understanding that next-generation technologies are new, complex, different in their performance, and known to introduce errors in reading (Hudson et al., 2014). This complexity has contributed, in part, to a long-standing uncertain regulatory and reimbursement environment for in vitro devices that has stymied both investment and momentum in diagnostic technology for some time (Hayes et al., 2013). Furthermore, although the clinical trial requirements for pharmaceuticals is understood and followed for all new drugs, an interesting situation has occurred with next-generation sequencing 1 The authors are participants in the activities of the IOM Roundtable on Translating Genomic-Based Research for Health. 2 Suggested citation: McCormack, R., and P. Billings. 2015. Clinical utility: Informing treatment decisions by changing the paradigm. Discussion Paper, Institute of Medicine, Washington, DC. http://www.iom.edu/ClinicalUtility. 1 in cancer. Clinicians quickly realized that driver mutations identified by next-generation sequencing are not unique to a specific cancer type, thus opening up the potential for off-label use of the targeted therapeutic. For example, drugs approved safe and effective for use with a specific mutation in one cancer, such as breast, could be equally safe and effective in other cancers, such as prostate, harboring the very same mutation. Unfortunately, these situations are not necessarily “quick wins." Even though the leap for an approved targeted drug that has demonstrated safety and effectiveness in one cancer to another cancer might be considered less risky than using a drug without any evidence of effectiveness and unknown safety, much of the same lengthy and expensive trials to demonstrate effectiveness in the off-label indication are required by regulators, payers, guideline developers, and providers before it is readily accepted as standard of care. Collectively, it is becoming clear that our ability to develop new technology and acquire potentially breakthrough information to benefit patients has exceeded our ability to readily reduce that information to practice within the confines of the existing clinical, regulatory, and reimbursement practices or requirements. The common denominator for much of this difficulty is insufficient evidence at all levels, from the laboratory test, to the laboratories conducting the testing, and to the repurposing of drugs for off-label use against a biomarker previously shown to be susceptible to that drug. For laboratory tests not developed as a companion diagnostic, the unequivocal demonstration of clinical utility has become the main requirement for a test to be covered by payers. Simply stated, clinical utility is defined as the use of a clinical test’s result to make a treatment decision that positively changes the outcome of a patient (Teutsch et al., 2009). This level of evidence represents a substantial challenge to all test developers and laboratories. Such studies are long and represent a human and financial commitment beyond the means of many companies. In addition, even if reimbursement is achieved, it usually represents a fraction of the company’s commitment to develop the technology, making such an investment unattractive (Hayes et al., 2013). However, recent efforts from all stakeholders to remedy the situation have resulted in progress across most of these barriers, especially as they pertain to next-generation technologies. Specifically, FDA guidance to standardize the level of performance across device manufacturers, technologies, and laboratories culminated in renewed regulatory direction (FDA, 2014). Although debatable, this guidance is aimed at obtaining a common ground and level of evidence for all device entities to achieve and move forward more confidently. Moreover, creative approaches to clinical studies have been implemented, such as stratifying clinical trials that use participant selection and biomarkers to enable rapid assessment of drug performance requiring fewer enrollees and, in many instances, less time (Tajik et al., 2013). This was complemented by efforts to create large databases as repositories for all biology and treatment decision results that could translate what was learned to a much wider audience of physicians struggling with the same problem, closing information gaps and guiding treatment decisions with confidence (Schilsky et al., 2014). These efforts will greatly accelerate clinical studies into testing these potentially new markers and acquire much-needed evidence and confidence supporting their use. The most recent opportunity to overcome these barriers comes in off-label use of currently approved targeted therapies. The proposed concept recognizes that relevant evidence on the drug’s safety and effectiveness exists and could translate to other cancers and patients but that exceptions do exist and patients’ safety must take priority (Di Nicolantonio et al., 2008). This 2 recent proposal conceptualizes a clinical registry trial that tightly defines patient eligibility and reasonable response end points, captures all patients’ demographic and clinical information in a database for continual learning, and is administered by a national oversight board for objective decision making. The patient is monitored closely for safety and therapeutic benefit or failure, and costs and risks are shared by all stakeholders (Schilsky, 2014). A similar model has also been suggested that proposes data sharing and early reviews of outcomes for appropriateness, but with the addition of payer coverage of all labeled drugs (Billings and Shather, 2014). This proposed framework allows for the accumulation of requisite data to fulfill regulatory and payer requirements for both the drug and device, while offering potentially beneficial therapy to patients in a controlled-risk environment. Such a departure from standard regulatory, clinical, and reimbursement practices was many years in the making and is less radical when viewed as a part of the methodical approach to collecting sufficient evidence from the bench to the patient, thus assuring patient safety, potential benefit, and possible new device-drug indications, all developed in a new paradigm. It is most appropriate that new technologies such as next-generation sequencing that initiated this evolution of personalized medicine are also largely responsible for this new paradigm. Below are three key issues we believe should be addressed to facilitate making treatment decisions using genomic-based technologies: 1. Clinical utility is a necessary addition to the evidentiary requirements for new technologies, but processes to define and measure clinical utility must be identified. 2. New paradigms in which all stakeholders share the burden and benefits of demonstrating such a level of evidence are required to rapidly translate findings from discovery to patient benefit. 3. To accommodate potentially beneficial new technology, there is a need for regulatory and reimbursement practices to be forward thinking to define a clear path to approval and coverage, avoiding delays in implementation once appropriate levels of clinical utility evidence have been established. REFERENCES Aston, J. L., and C. Lovly. 2014. Anticancer agents. http://www.mycancergenome.org/content/ other/molecular-medicine/anticancer-agents/ (accessed November 10, 2014). Billings, P. R., and B. Shather. 2014. Off label cancer therapies (manuscript submitted). Di Nicolantonio, F., M. Martini, F. Molinari, A. Sartore-Bianchi, S. Arena, P. Saletti, S. De Dosso, L. Mazzucchelli, M. Frattini, S. Siena, and A. Bardelli. 2008. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. Journal of Clinical Oncology 26(35):5705–5712. FDA (Food and Drug Administration). 2014. Framework for regulatory oversight of laboratory developed tests (LDTs): Draft guidance. http://www.fda.gov/downloads/MedicalDevices/ DeviceRegulationandGuidance/GuidanceDocuments/UCM416685.pdf (accessed November 10, 2014). Hayes, D. F., J. Allen, C. Compton, G. Gustavsen, D. G. Leonard, R. McCormack, L. Newcomer, K. Pothier, D. Ransohoff, R. L. Schilsky, E. Sigal, S. E. Taube, and S. R. Tunis. 2013. Breaking a vicious cycle. Science Translational Medicine 5(196):196cm196. 3 Hudson, A. M., T. Yates, Y. Li, E. W. Trotter, S. Fawdar, P. Chapman, P. Lorigan, A. Biankin, C. J. Miller, and J. Brognard. 2014. Discrepancies in cancer genomic sequencing highlight opportunities for driver mutation discovery. Cancer Research 74(22):6390-6396. Schilsky, R. L. 2014. Implementing personalized cancer care. Nature Reviews Clinical Oncology 11(7):432–438. Schilsky, R. L., D. L. Michels, A. H. Kearbey, P. P. Yu, and C. A. Hudis. 2014. Building a rapid learning health care system for oncology: The regulatory framework of CancerLinQ. Journal of Clinical Oncology 32(22):2373–2379. Tajik, P., A. H. Zwinderman, B. W. Mol, and P. M. Bossuyt. 2013. Trial designs for personalizing cancer care: A systematic review and classification. Clinical Cancer Research 19(17):4578–4588. Teutsch, S. M., L. A. Bradley, G. E. Palomaki, J. E. Haddow, M. Piper, N. Calonge, W. D. Dotson, M. P. Douglas, and A. O. Berg. 2009. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) initiative: Methods of the EGAPP working group. Genetics in Medicine 11(1):3–14. 4
© Copyright 2026